Last updated on April 10, 2025
What if, you don’t have to worry about massive datasets? What if they’re open and easily accessible to you? Would it help… to inspire you to work on that bioinformatics project?
Fret not! Have you heard of AWS Open Data Program (ODP)? You will now!
Related: If you haven’t read about utilizing SRA Toolkit and AWS for data preparation, you can read it here.
Introduction to Related Concepts: Why Genomic Data in Amazon S3?
First of all, what is this AWS Open Data Program (ODP), and why does it store genomic data in Amazon S3?
On the official website of AWS, you can see in the Genomic Resources section that the following is written about the AWS Open Data Program (ODP):
“The AWS Open Data Program (ODP) helps democratize data access by making it readily available in Amazon S3, providing the research community with a single documented source of truth. For a complete list of the >40 genomics open data sets available, see The AWS Open Data Program (ODP).”
From this, we can note the following benefits:
- Data democratization: Since the data are uploaded publicly on AWS, anyone worldwide can access the necessary data.
- Cost efficiency: With the data uploaded on Amazon S3, you can directly work with it using AWS services, lowering the costs needed for analysis.
- Scalability and performance: You can also use AWS’s scalable nature since the data is stored in Amazon S3. One example is the integration of NIH’s Sequence Read Archive (SRA) into ODP. Again, if you are curious about how to work with SRA and AWS for your data preparation, you can check it out here.
- Centralized data repository: The data you need is now stored in a single repository, which is the ODP. Having a centralized place ensures data consistency and reliability.
Open Data on AWS
Let’s dig into this further. When we say “Open data on AWS,” it means that the data is shared on AWS, right? What does that mean for us? This means anyone can analyze the data and build services on top of it with the help of several AWS services such as Amazon EC2, Amazon Athena, AWS Lambda, and Amazon EMR.
AWS Data Exchange
For easier searching of publicly made available datasets through AWS services, AWS Data Exchange is there for you. You can access it here.
Open Data Sponsorship Program
You are familiar with the fact that when using AWS resources, there has to be some payment happening to maintain its usage. AWS works with data providers who wish to share their datasets, but there are terms and conditions in place before they can share. Then, AWS will cover the storage cost of that data shared for public use.
Going back, you can check out here for a complete list of resources available for your genomic workflows.
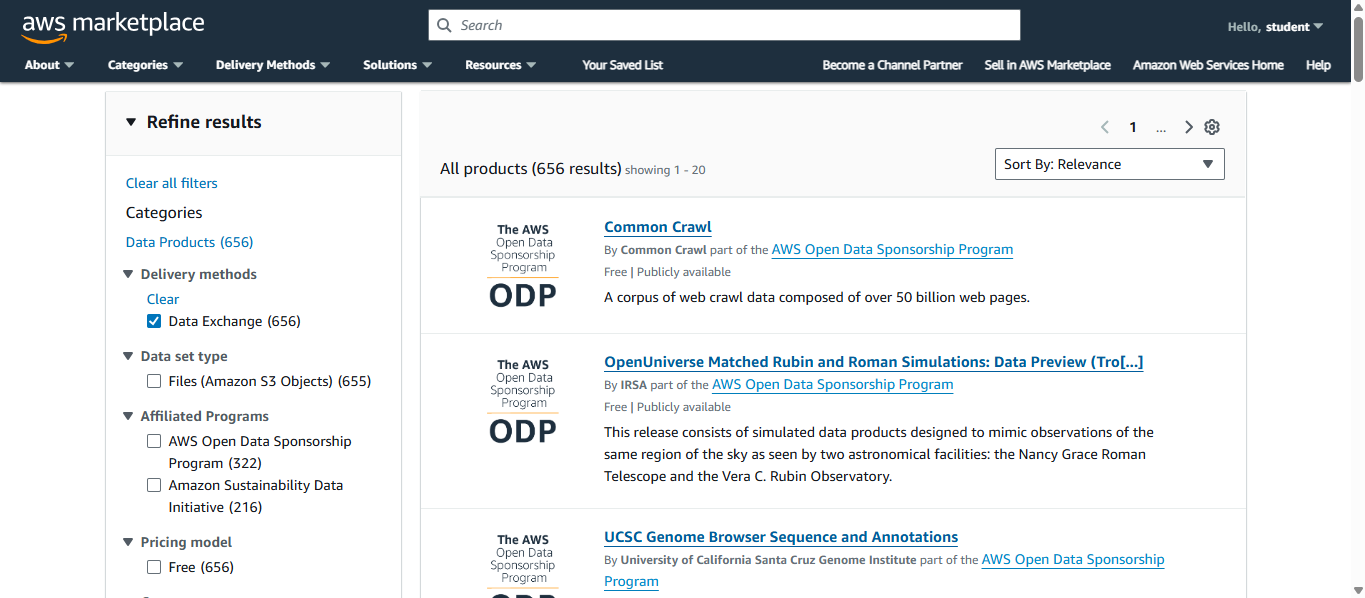
Below are some of the genomic data that you can access and that is available in ODP.
Later, we will be looking into them for examples!
Getting Started with Genomic Data on Amazon S3
Let’s go back quickly to Amazon S3 and how it is a good storage solution in general. Amazon S3 is a scalable and durable object storage, meaning it can handle the increase of your data so you wouldn’t need to worry about meticulously planning for it before doing anything. Your data is also stored across multiple devices in multiple facilities within an AWS Region.
Now, we can explore the different examples of datasets in OPD!
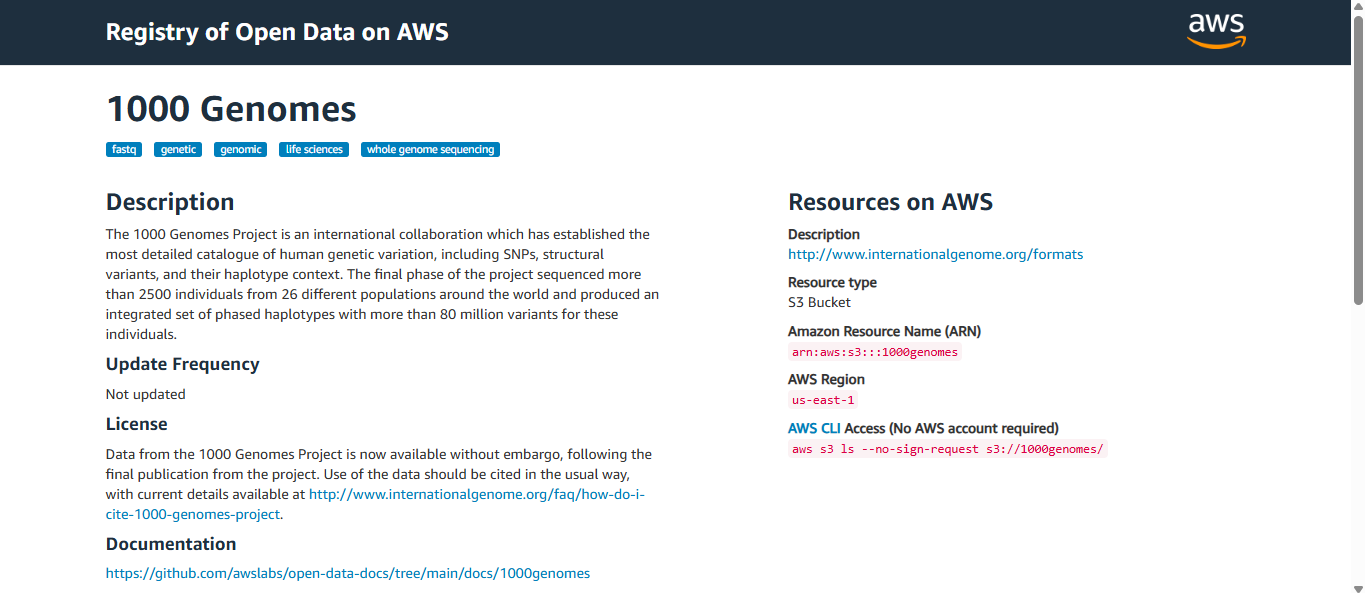
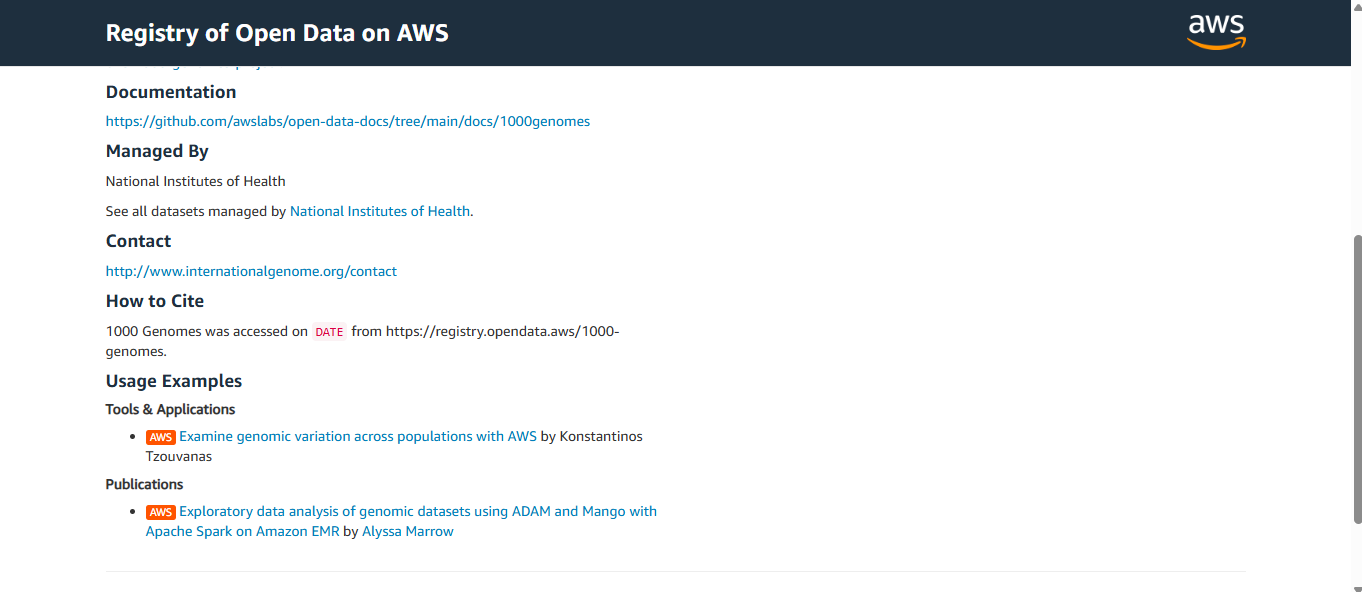
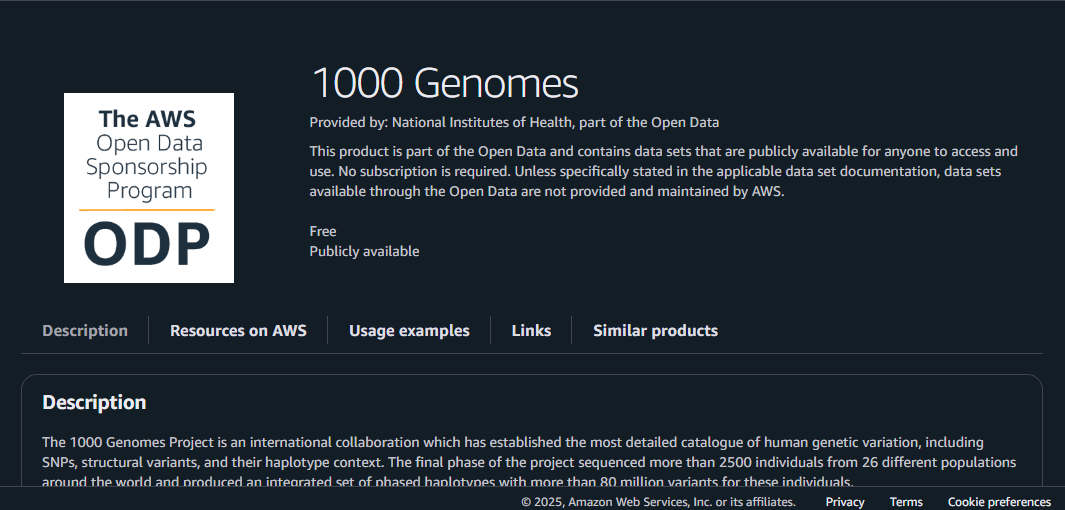
If you click on one of them (in this case, the one titled “1000 Genomes”), you will be greeted by the necessary information you need about it.
Below is a picture for reference.
Let’s go through the parts one by one! On the left side…
- “1000 Genomes” – This is the name of the dataset.
- Tags – Those keywords with the blue background are tags related to this dataset. It helps users like you to find relevant datasets quickly. In our example:
- fastq – format of raw sequencing data
- genetic/genomic – relating to genome
- whole genomic sequencing – type of data
- Description – Explains the background of the dataset, what it contains, and its significance.
- Update Frequency – Tells you how regularly updated this dataset is.
- License – Let’s you know about the proper use of this dataset (citation policy and licensing terms).
- Documentation – A link to GitHub documentation will help you throughout the use of this dataset.
- Managed By – Who manages the dataset? In this case, it’s the National Institutes of Health.
- Contact – Who to contact regarding the dataset.
- How to Cite – A guide for proper citation of the dataset.
- Usage Examples – This is to help you understand the dataset you’re working with.
On the right side is the AWS-related information you need to use. We’ll go through a short hands-on to understand this fully.
Implementation Steps
Now it’s your turn to try it out! Let’s get the steps down first, shall we?
Prerequisites
- Have an AWS account to access the services involved.
- Have AWS CLI installed. You can check it out here in this article.
Finding datasets
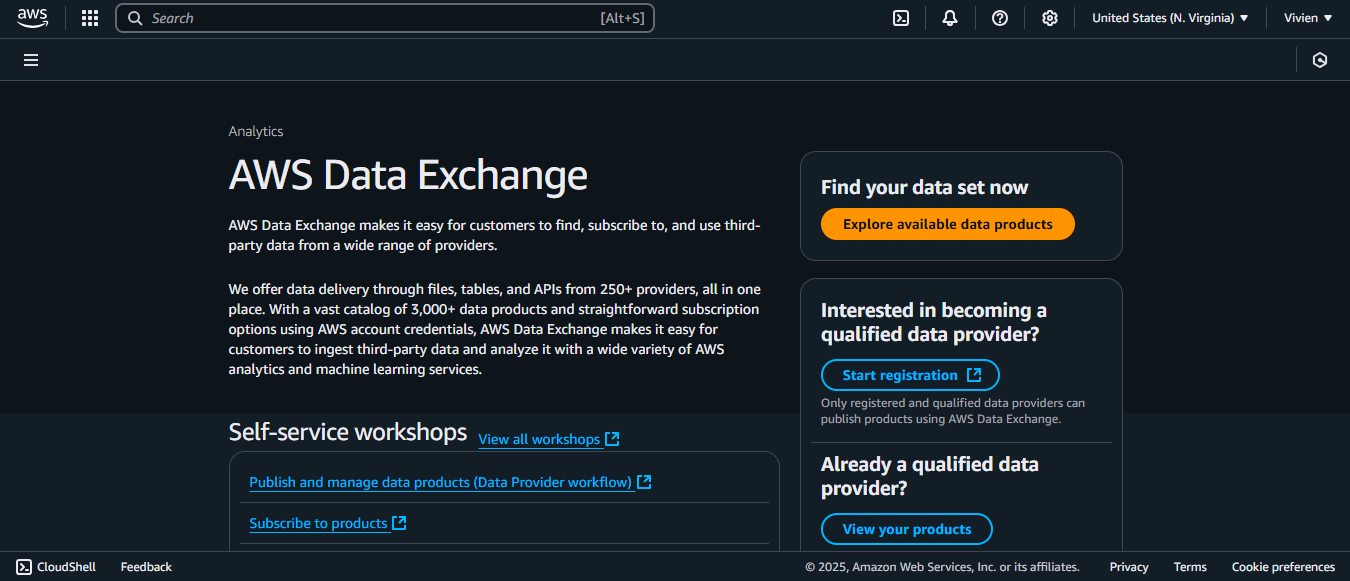
1. First, navigate to the AWS Data Exchange console.
2. You can click the “Explore available data products” button.
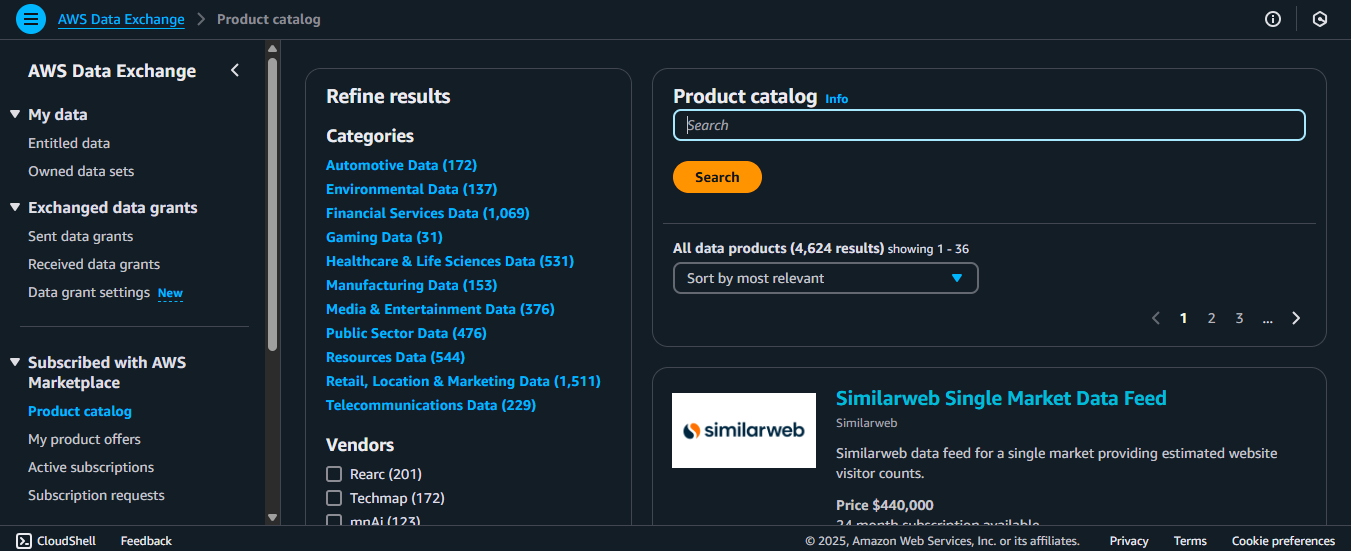
3. As you can see, the different parts are here.
- There’s the left panel for quick access to different options within this service.
- You can refine the dataset results by category or vendor.
- You can also directly search for the dataset on the far right.
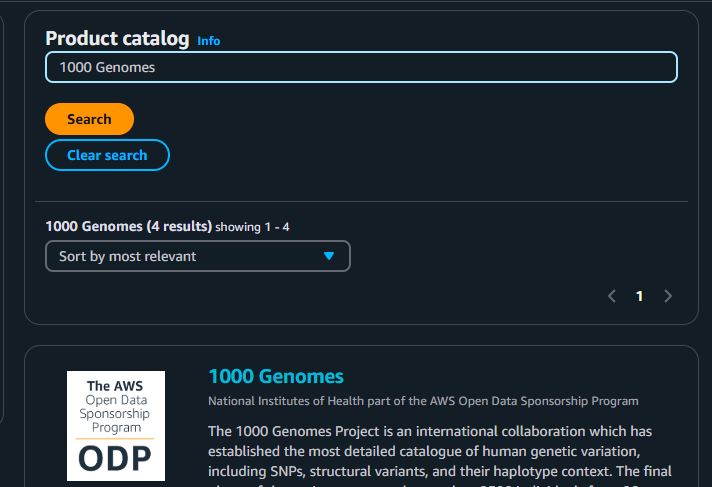
4. For this example, we will use the “1000 Genomes” dataset you saw earlier. So type that one in!
5. You can see the following once you click the “1000 Genomes” dataset. It will seem a bit different from where we have viewed this dataset, but it’s just the same.
Accessing data
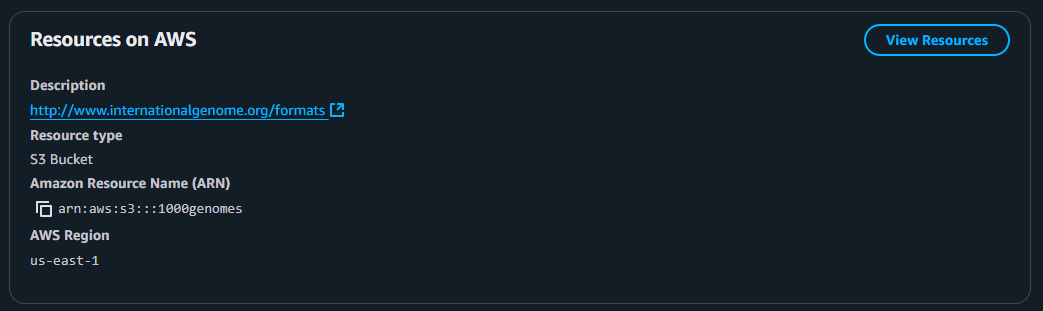
1. Navigate to the “Resources on AWS” tab. These are the information you want to be able to access the dataset.
2. Make sure you have downloaded AWS CLI in your computer and did the necessary configurations. Let’s focus on the Amazon Resource Name (ARN) of this dataset: arn:aws:s3:::1000genomes. This means…
arn:aws:s3:::– tells you it’s an S3 bucket1000genomes– the name of the bucket
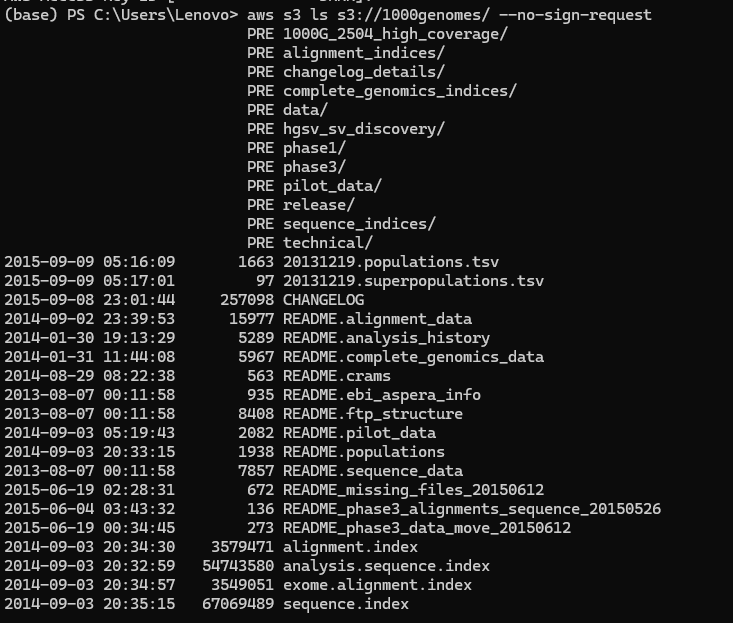
3. Enter this command on your command line. This will enable you to first list the contents this S3 bucket has. You will use the name of the bucket to access it!
aws s3 ls s3://1000genomes/ --no-sign-request 
4. You can also explore inside a folder with this command:
aws s3 ls s3://1000genomes/phase3/ --no-sign-request
5. Finally, of course, you can download a specific file from this dataset. You will be downloading a BAM file for this exercise. (Want to know more about FASTQ files and other file types? Check it out here).
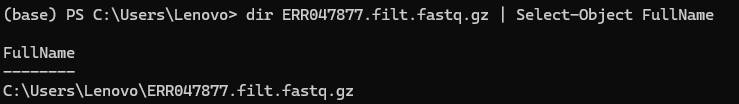
aws s3 cp s3://1000genomes/phase3/data/NA21144/sequence_read/ERR047877.filt.fastq.gz ./ --no-sign-request
6. Now it’s downloaded on your computer! You can input this command to get its complete file location and access it.
You have successfully accessed one of the publicly available data in AWS ODP! Great work!
Conclusion: Empowering Genomic Research with Amazon S3
In this article, you’ve learned and understood how AWS Open Data Program enables you to access publicly available data. With Amazon S3 where those data are stored, you can directly use AWS services on the said data. Also, you don’t have to take too long to search for datasets if they are already there, lessening the time and effort needed for data storage and management.
References
Amazon Open Data Sponsorship Program
Amazon S3 – Cloud Object Storage – AWS
https://tutorialsdojo.com/commanding-genomics-aws-cli-for-bioinformatics-workflows/
https://tutorialsdojo.com/biology-data-the-cloud-aws-in-bioinformatics-awshealthomics/