Last updated on March 24, 2025
“This is becoming more troublesome…” I can hear you mumble as you move your mouse to click on the different AWS services you need in the console.
As time goes by, it can get tiresome to keep on clicking things for your workflow instead of doing them in just one place. You can streamline this process with the AWS CLI! Knowing not only how to work with AWS Console, but also the AWS Command-Line Interface will help you up your bioinformatics workflow!
What is AWS CLI?
According to their official website,
“The AWS Command Line Interface (AWS CLI) is an open source tool that enables you to interact with AWS services using commands in your command-line shell. With minimal configuration, the AWS CLI enables you to start running commands that implement functionality equivalent to that provided by the browser-based AWS Management Console from the command prompt in your terminal program.”
You can use these commands on the following:
- On your local computer – either in Linux/MacOS or command prompt/PowerShell (Windows)
- On remote servers – such as Amazon EC2 instances with tools like SSH or PuTTY
- In managed AWS services – using the CLI with AWS Systems Manager for running commands
Currently, the most recent version is the AWS CLI version 2. If you are already using the AWS CLI version 1, see here about the migration process, otherwise, you can proceed as written in this article.
Getting Started
1. Installation and Configuration
The blog will show Windows as an example, but it also works with other operating systems. Kindly refer to the installation guide of AWS.
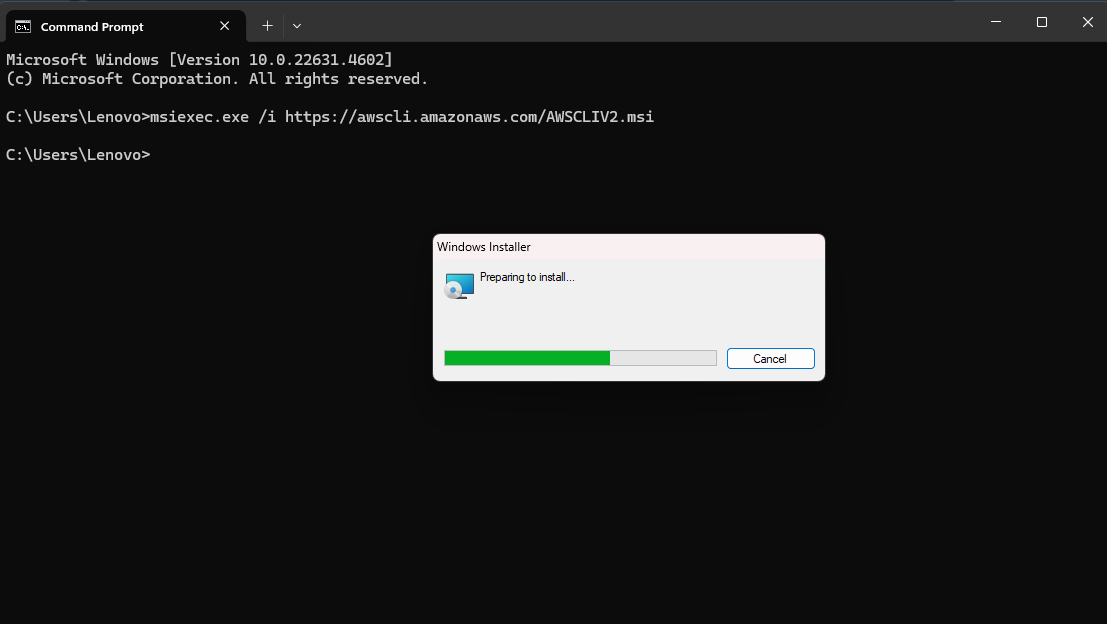
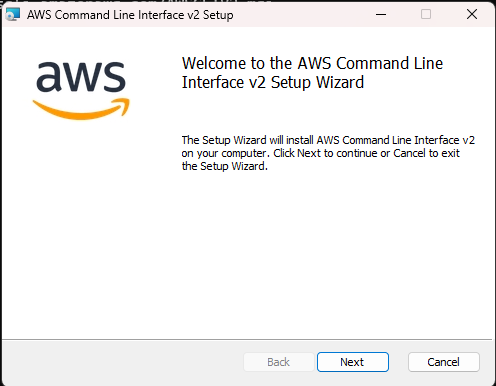
- Download the AWS CLI installer compatible with your operating system (Installation Guide). Follow the corresponding steps indicated in the article.
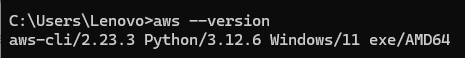
- Verify installation by running the command: aws –version in your command line.
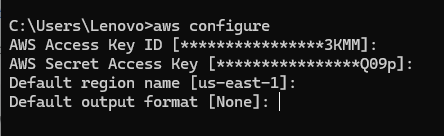
- Configure AWS CLI with your credentials: aws configure
Then, you’ll be prompted to input:
- Access Key ID
- Secret Access Key
- Default region (e.g., us-west-2)
- Output format (e.g., json, text, or table)
The Access Key ID and Secret Access Key are created after you have set up your account and configured the IAM roles and users.
2. Setting up your CLI for your projects
It is important to set up your CLI properly for more efficient workflows. Do the same as in the previous number and take note of the following:
- Before starting your workflow, set up the users and roles necessary. Make it a habit to not work with your root account for added security.
- To ensure low latency and cost efficiency, set the region to where your data is being processed and analyzed.
Related Terminologies
Before diving into the different CLI commands, you can familiarize yourself with the following terminologies:
- Sequence store – store genomic sequence data
- Reference store – holds your reference genome files
- Variant store – for genomic variant data used in analysis
- Annotation store – for more information about those variants
- Job – a specific task or operation done in AWS environment
- Activation job – the process of restoring archived read sets making them ready for analysis
- Read sets – collection of genomic sequence reads (FASTQ, BAM, CRAM)
- Workflow – series of steps or tasks for processing and analyzing genomic data
- Run – single execution of a workflow
- Run group – a collection of workflow runs sharing certain configurations
- Workflow run – execution instance of a workflow
You can also check out my other bioinformatics articles for more information here!
Key AWS CLI Commands for Bioinformatics
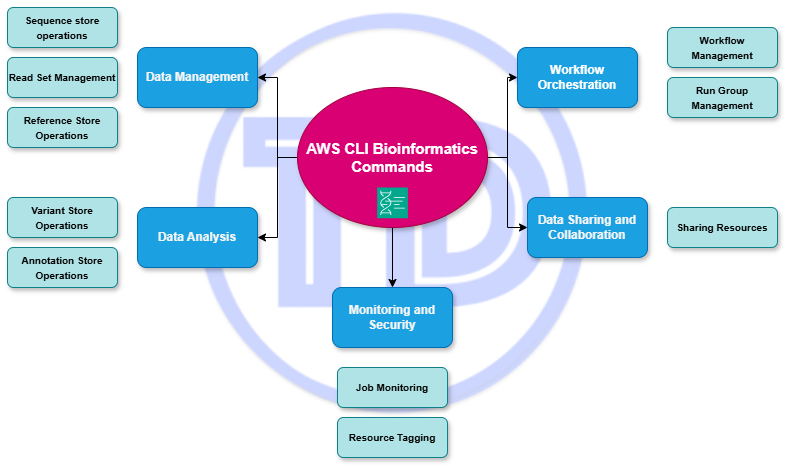
Listed below are the common AWS CLI commands that you can reference for your bioinformatics workflow. It is categorized into five (5): data management, workflow orchestration, data analysis, data sharing and collaboration, and monitoring and security.
Take note that the commands are not limited to these. You can check the documentation here for more details.
Data Management
|
Sequence store operations |
|
|
Command |
Description |
|
aws omics create-sequence-store \ –name my-seq-store |
Create a sequence store |
|
aws omics delete-sequence-store \ –id 1234567890 |
Delete a sequence store |
|
aws omics list-sequence-stores |
List sequence stores |
|
aws omics get-sequence-store \ –id 1234567890 |
Get sequence store details |
|
Read Set Management |
|
|
aws omics get-read-set-import-job \ –sequence-store-id 1234567890 \ –id 1234567890 |
View read set import job |
|
aws omics get-read-set-export-job \ –sequence-store-id 1234567890 \ –id 1234567890 |
View read sets export job |
|
aws omics batch-delete-read-set \ –sequence-store-id 1234567890 \ –ids 1234567890 0123456789 |
Delete multiple read sets |
|
Reference Store Operations |
|
|
aws omics create-reference-store \ –name my-ref-store |
Create a Reference Store |
|
aws omics delete-reference-store \ –id 1234567890 |
Delete a Reference Store |
|
aws omics start-reference-import-job \ –reference-store-id 1234567890 \ –role-arn arn:aws:iam::123456789012:role/omics-service-role-serviceRole-W8O1XMPL7QZ \ –sources sourceFile=s3://omics-artifacts-01d6xmpl4e72dd32/Homo_sapiens_assembly38.fasta,name=assembly-38 |
Import References |
|
aws omics get-reference-store \ –id 1234567890 |
View a reference store |
Workflow Orchestration
|
Workflow Management |
|
|
aws omics create-workflow \ –name cram-converter \ –engine WDL \ –definition-zip fileb://workflow-crambam.zip \ –parameter-template file://workflow-params.json |
Create a workflow |
|
aws omics start-run \ –workflow-id 1234567 \ –role-arn arn:aws:iam::123456789012:role/omics- service-role-serviceRole-W8O1XMPL7QZ \ –name ‘cram-to-bam’ \ –output-uri s3://omics-artifacts- 01d6xmpl4e72dd32/workflow-output/ \ –run-group-id 1234567 \ –priority 1 \ –storage-capacity 10 \ –log-level ALL \ –parameters file://workflow-inputs.json |
Start a workflow run |
|
aws omics delete-workflow \ –id 1234567 |
Delete workflow |
|
aws omics get-workflow \ –id 1234567 |
Get workflow details |
|
aws omics list-workflows |
List workflows |
|
Run Group Management |
|
|
aws omics create-run-group \ –name cram-converter \ –max-cpus 20 \ –max-duration 600 |
Create a run group |
|
aws omics delete-run-group \ –id 1234567 |
Delete a run group |
|
aws omics get-run-group \ –id 1234567 |
Get run group details |
Data Analysis
|
Variant Store Operations |
|
|
aws omics create-variant-store \ –name my_var_store \ –reference referenceArn=arn:aws:omics:us-west-2:123456789012:referenceStore/1234567890/reference/1234567890 |
Create a variant store |
|
aws omics delete-variant-store \ –name my_var_store |
Delete a variant store |
|
aws omics start-variant-import-job \ –destination-name my_var_store \ –no-run-left-normalization \ –role-arn arn:aws:iam::123456789012:role/omics-service-role-serviceRole-W8O1XMPL7QZ \ –items source=s3://omics-artifacts-01d6xmpl4e72dd32/Homo_sapiens_assembly38.known_indels.vcf.gz |
Start variant import job |
|
aws omics cancel-variant-import-job \ –job-id 69cb65d6-xmpl-4a4a-9025-4565794b684e |
Cancel variant import job |
|
Annotation Store Operations |
|
|
aws omics create-annotation-store \ –name my_ann_store \ –store-format VCF \ –reference referenceArn=arn:aws:omics:us-west-2:123456789012:referenceStore/1234567890/reference/1234567890 |
Create an annotation store |
|
aws omics delete-annotation-store \ –name my_vcf_store |
Delete an annotation store |
|
aws omics start-annotation-import-job \ –destination-name tsv_ann_store \ –no-run-left-normalization \ –role-arn arn:aws:iam::123456789012:role/omics-service-role-serviceRole-W8O1XMPL7QZ \ –items source=s3://omics-artifacts-01d6xmpl4e72dd32/targetedregions.bed.gz |
Start annotation import job |
|
aws omics cancel-annotation-import-job \ –job-id 04f57618-xmpl-4fd0-9349-e5a85aefb997 |
Cancel annotation import job |
Data Sharing and Collaboration
|
Sharing Resources |
|
|
aws omics create-share \ –resource-arn “arn:aws:omics:us-west-2:555555555555:variantStore/omics_dev_var_store” \ –principal-subscriber “123456789012” \ –name “my_Share-123” |
Create a share |
|
aws omics accept-share \ —-share-id “495c21bedc889d07d0ab69d710a6841e-dd75ab7a1a9c384fa848b5bd8e5a7e0a” |
Accept a share |
|
aws omics delete-share \ –share-id “495c21bedc889d07d0ab69d710a6841e-dd75ab7a1a9c384fa848b5bd8e5a7e0a” |
Delete a share |
|
aws omics list-shares \ –resource-owner SELF |
List shares |
Monitoring and Security
|
Job Monitoring |
|
|
aws omics get-read-set-activation-job \ –sequence-store-id 1234567890 \ –id 1234567890 |
View archived read set activation job |
|
aws omics list-read-set-activation-jobs \ –sequence-store-id 1234567890 |
List read set activation jobs |
|
aws omics get-run \ –id 1234567 |
Get run details |
|
aws omics list-runs |
List runs |
|
Resource Tagging |
|
|
aws omics tag-resource \ –resource-arn arn:aws:omics:us-west-2:123456789012:workflow/1234567 \ –tags department=analytics |
Tag a resource |
|
aws omics untag-resource \ –resource-arn arn:aws:omics:us-west-2:123456789012:workflow/1234567 \ –tag-keys department |
Untag a resource |
|
aws omics list-tags-for-resource \ –resource-arn arn:aws:omics:us-west-2:123456789012:workflow/1234567 |
List tags for a resource |
Conclusion
Now you know how you can utilize the AWS CLI for your bioinformatics workflow! You are encouraged to explore the CLI and experiment with your projects. With this new skill, you will be able to work efficiently by streamlining your process!
References
All about AWS CLI
What is the AWS Command Line Interface? – AWS Command Line Interface
Migrating from AWS CLI version 1 to AWS CLI version 2 – AWS Command Line Interface
AWS CLI Commands in Bioinformatics
My other articles
AWS in Bioinformatics: Biology, data, & the cloud
SRA Toolkit + AWS: Revolutionizing Bioinformatics Data Prep
Unlocking the Secrets of DNA: Analyzing Genomic Data with AWS